Proteins from junk DNA
The complexity of living organisms has increased over the last 500 million years, and yet the number of known genes has remained constant. It is also a conundrum that only 1.5% of the human genome is made up of known genes and the remaining 98.5% is unknown. And there are many other questions that nag scientists: Do living organisms make new genes? If they do, how do these novel genes emerge? Do novel genes emerge by accident, or do they emerge with a purpose-built function? Can the novel proteins encoded by novel genes form structures? How do the novel genes drive evolution? Are novel proteins disrupted in diseases?
My research focuses on novel genes in different organisms, and I investigate the biological functions of the novel proteins they encode to determine whether they are disrupted in diseases. Four of my lab's papers were published in the last few weeks and we are excited to share the results and their implications with a wider community. The novel genomic regions I investigate cannot be defined by our current definition of a gene; hence, we call these novel regions "novel open reading frames," or nORFs.
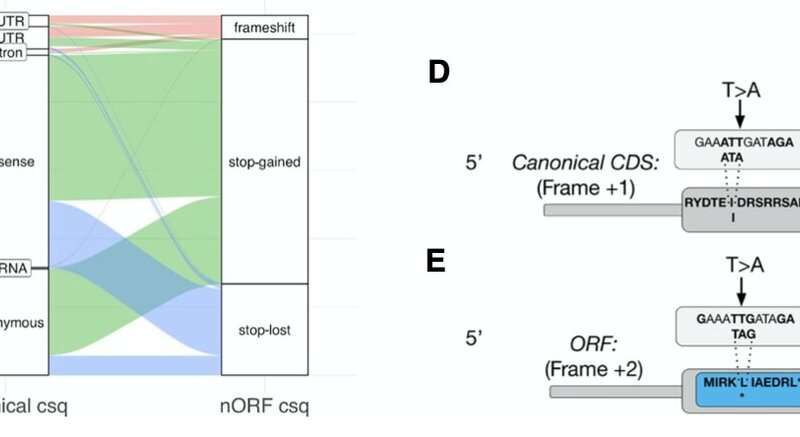
In Neville et al., 2021, we cataloged, curated, annotated and released nORFs in the human genome. We show that thousands of variants that are classified as benign have to be reclassified based on their consequence to novel proteins (Fig. 1). In Erady et al., 2021, we show that novel proteins can form structures that undergo biochemical regulation like known proteins and be targeted by drugs (Fig. 2). In Puntambekar et al., 2020, we investigated the speciation of cichlid fishes and show that novel genes can evolve faster and can potentially explain the speciation processes (Fig. 3). In Gunnarsson and Prabakaran, 2021, we show that novel genes are expressed in the malarial parasite Plasmodium falciparum. Some are expressed only when the parasite infects mosquitoes and some are expressed when they infect humans (Fig. 4). These observations open the door for new drug targets and possible cures for malaria.
-

Fig. 2 Credit: Erady, C. et al., 2021. Pan-cancer analysis of transcripts encoding novel open-reading frame (nORFs) and their potential biological functions. NPJ genomic medicine, 6(1), p.4. -
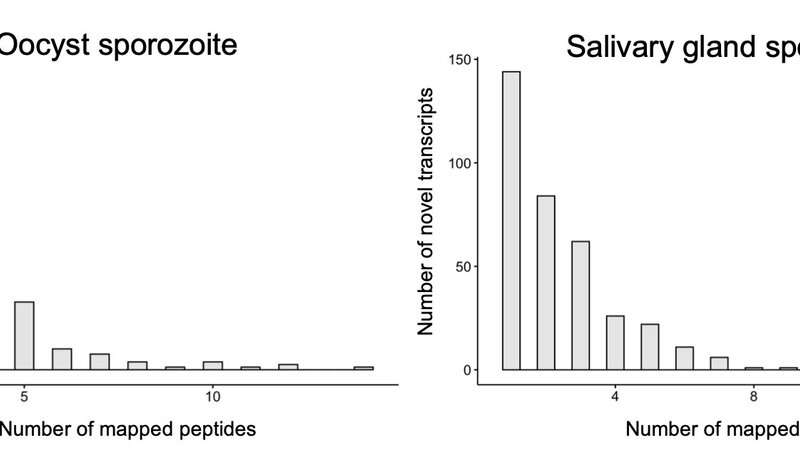
Fig. 3. Credit: Gunnarsson, S., and Prabakaran, S., 2021. In silico identification of novel open reading frames in Plasmodium falciparum oocyte and salivary gland sporozoites using proteogenomics framework. To appear in Malaria Journal
These results demonstrate that under stress, physiological changes, or adaptive radiation, nature tinkers with the evolvable noncoding genome to make new parts as and when needed. We speculate that all known genes are just for housekeeping while the novel genes are the ones that perform specific functions. This calls for the redefinition of a gene and change in our understanding of evolution.
This story is part of Science X Dialog, where researchers can report findings from their published research articles. Visit this page for information about ScienceX Dialog and how to participate.
More information: Neville, M.D.C. et al., 2021. A platform for curated products from novel open reading frames prompts reinterpretation of disease variants. Genome research. Available at: dx.doi.org/10.1101/gr.263202.120
Erady, C. et al., 2021. Pan-cancer analysis of transcripts encoding novel open-reading frame (nORFs) and their potential biological functions. NPJ genomic medicine, 6(1), p.4.
Puntambekar, S. et al., 2020. Evolutionary divergence of novel open reading frames in cichlids speciation. Scientific reports, 10(1), p.21570.
Gunnarsson, S., and Prabakaran, S., 2021. In silico identification of novel open reading frames in Plasmodium falciparum oocyte and salivary gland sporozoites using proteogenomics framework. To appear in Malaria Journal
Sudhakaran Prabakaran is a Lecturer (Assistant Professor) in the Department of Genetics at the University of Cambridge, UK. He is also a Fellow of St. Edmund's College, University of Cambridge, UK, and co-founder of NonExomics, LLC.